If you’re visiting this website, it’s likely that you have some microscopy data that you want to use. Still, it’s good to start working with some example data for the following reason. If something from this website doesn’t work, there can be two reasons why:
- the method doesn’t work.
- the method works, but not for your data.
To exclude reason 2, we work with example microscopy data. We use the following example datasets:
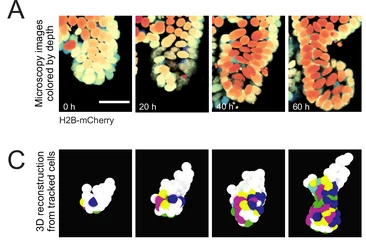
Intestinal organoid crypts
Intestinal organoids were the first organoids. They have protrusions called crypts. In this dataset, nine of such crypts were manually tracked. The rest of the organoid was not tracked.
From the paper: Huelsz-Prince, G. et al. Mother cells control daughter cell proliferation in intestinal organoids to minimize proliferation fluctuations. eLife 11, e80682 (2022). https://doi.org/10.7554/eLife.80682
C. Elegans developing embryos
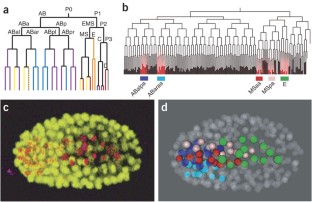
Not an organoid, but a worm embryo. Still, the data looks a lot like data you would expect in organoids. The training dataset includes two worms, the validation dataset includes a third worm.
Download on this page (scroll down a bit on that page)
From the paper: Murray, J. et al. Automated analysis of embryonic gene expression with cellular resolution in C. elegans. Nature Methods 5, 703–709 (2008). https://doi.org/10.1038/nmeth.1228
Know another dataset?
Do you have other 3D+time datasets that could be useful for demonstrating cell tracking? Please let me know, then I can add them to this page!